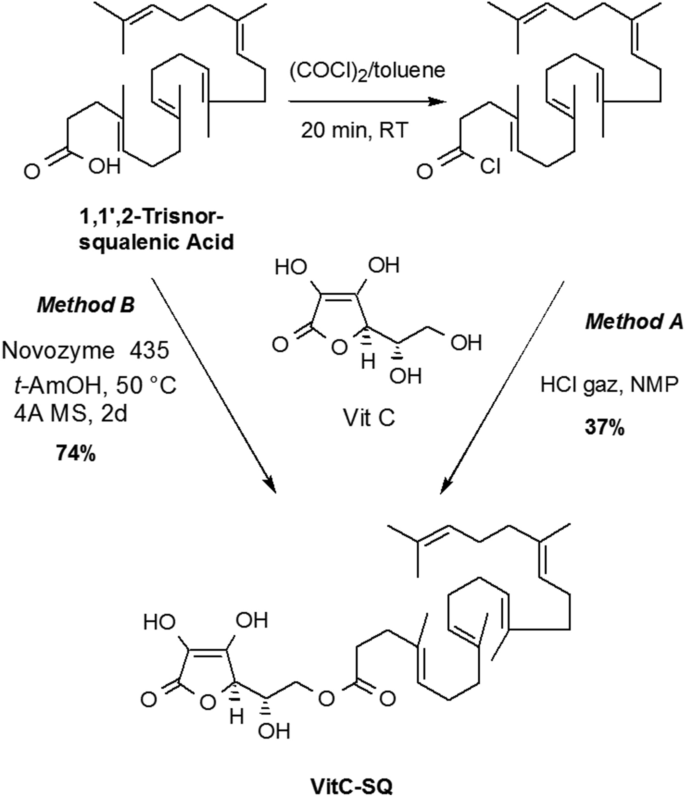
Synthesis of Vit C–SQ
IR spectra were obtained as solid or neat liquid on a Fourier Transform Bruker Vector 22 spectrometer. Only significant absorptions are listed. The 1H and 13C NMR spectra were recorded on Bruker Avance 300 (300 MHz and 75 MHz, for 1H and 13C, respectively) or Bruker Avance 400 (400 MHz and 100 MHz, for 1H and 13C, respectively) spectrometers. Recognition of methyl, methylene, methine, and quaternary carbon nuclei in 13C NMR spectra rests on the J-modulated spin-echo sequence. Mass spectra were recorded on a Bruker Esquire-LC. Analytical thin-layer chromatography was performed on Merck silica gel 60F254 glass precoated plates (0.25 mm layer). Column chromatography was performed on Merck silica gel 60 (230–400 mesh ASTM). Toluene and N-methyl pyrrolidone (NMP) were distilled from calcium hydride, under a nitrogen atmosphere. t-AmOH, was dried of sodium and distilled. All reactions involving air- or water-sensitive compounds were routinely conducted in glassware which was flame-dried under a positive pressure of nitrogen or argon. Ascorbic acid, oxalyl chloride, squalene, NMP and Novozyme 435 (L4777) were purchased from Sigma-Aldrich Chemical Co., France. Chemicals obtained from commercial suppliers were used without further purification. 1,1′,2-tris-norsqualenic acid was prepare according to Ceruti et al.32. For comparison purposes, palmitoyl ascorbate was prepared from palmitic acid and ascorbic acid according to litterature54.
4,8,13,17,21-Pentamethyl-docosa-4,8,12,16,20-pentaenoyl chloride (2)
A solution of trisnorsqualenic acid (1.80, 4.5 mmol) in anhydrous toluene (10 mL) was degassed by bubbling a stream of nitrogen through the solution for 5 min. Oxalyl chloride (1.74 g, 13.8 mmol) was added dropwise at 20 °C. The reaction mixture was stirred for 3 h at the same temperature and concentrated under reduced pressure to give the title compound as a yellow oil which is used directly in the next step. IR (film) ν: 2920, 1799, 1443, 1382, 955, 893 cm−1; RMN 1H (300 MHz, CDCl3) δ : 5.23–5.10 (m, 5 H), 2.45 (t, J = 7.3 Hz, 2 H), 2.30 (t, J = 7.5 Hz, 2H), 2.15–1.95 (m, 16 H), 1.70 (s, 3H), 1.62 (s, 15H) ppm; RMN 13C (75 MHz, CDCl3), δ :173.2 (C, COCl), 135.1 (C, C=CH), 134.8 (C, C=CH), 134.6 (C, C=CH), 131.4 (C, C=CH), 131.1 (C, C=CH), 126.6 (CH, C=CH), 124.7 (CH, C=CH), 124.4 (CH, C=CH), 124.3 (2CH, C=CH), 45.8 (CH2), 39.7 (2CH2), 34.4 (CH2), 34.6 (CH2), 28.2 (2CH2), 26.8 (CH2), 26.7 (CH2), 26.5 (CH2), 25.7 (CH3, C=C(CH3)2), 17.6 (CH3), 16.0 (CH3), 15.9 (2 CH3), 15.8 (CH3) ppm.
6-O-[4,8,13,17,21-Pentamethyl-docosa-4,8,12,16,20-pentaenoyl] ascorbic acid, method A
A stream of dry HCl was passed through 5 mL of N-methylpiperidone in a washing bottle until the entire mass solidified. N-methylpyrrolidinone was added to get a clear solution (~ 5 mL) which was transferred into a 50 mL round bottom flask. Ascorbic acid (0.7 g, 4.0 mmol) was added and the mixture was stirred until complete dissolution. The reaction mixture was cooled to 0 °C and squalenoyl chloride (418 mg, 1.0 mmol) was added. After being stirred for 20 h at room temperature, water was added (20 mL) and the mixture was extracted with ethyl acetate (4 × 20 mL). The combined organic layers were washed with water (2 × 5 mL) dried over MgSO4 and concentrated under reduced pressure. The residual NMP was removed under reduced pressure using dry ice rotavapor (60 °C, 0.05 mm Hg,) to leave an oil which was purified by chromatography over silica gel eluting with ethyl acetate and then AcOEt/MeOH (98:2) to give the title compound as a pale yellow thick oil (190 mg, 34%).
6-O-[4,8,13,17,21-Pentamethyl-docosa-4,8,12,16,20-pentaenoyl] ascorbic acid, method B
To a suspension of ascorbic acid (200 mg, 2 mmol) in anhydrous tert-amyl alcohol (5 mL) was successively added Novozyme 435 (150 mg), tris-norsqualenic acid (200 mg, 1.0 mmol) and 1 g of 4 Å molecular sieves. The flask was fixed to a rotavapor and slowly stirred at 50 °C (water bath). After 48 h, the reaction mixture was cooled and filtered through a plug of Celite. The solid was thoroughly washed with AcOEt and the filtrate was concentrated under reduced pressure. The crude product was purified by chromatography over silica gel eluting with ethyl acetate and then AcOEt/MeOH (98:2) to give the title compound as a pale yellow thick oil (209 mg, 74%); [α]D = + 10 (c = 0.5, EtOH); IR (film) ν: 3600–2800, 2976, 2857, 1744, 1696, 1443, 1382, 1350, 1296, 1151, 1119, 1043, 983, 844 cm−1; NMR 1H (360 MHz, MeOH-d4) δ: 5.20–5.05 (m, 5H), 4.72 (d, J = 2.0 Hz, 1H, H-4), 4.24 (dd, J = 11.16 Hz , J = 6.84 Hz, 1H, H-6), 4.18 (dd, J = 11.16 Hz , J = 6.12 Hz, 1H, H-6), 4.08 (ddd, J = 6.84 Hz, J = 6.12 Hz, J = 2.0 Hz, 1H, H-5), 2.46 (t, J = 7.74 Hz, 2H, COCH 2CH2), 2.30 (t, J = 7.56 Hz, 2H, COCH2CH 2), 2.12–1.92 (m, 16H), 1.63 (d, J = 1.08 Hz, 3H, HC = C(CH 3)2), 1.62 (s, 3H), 1.60 (s, 12H) ppm; NMR 13C (90.5 MHz, CDCl3) δ: 174.6 (C, CO), 173.1 (C, C1), 154.0 (C, C3), 136.0 (C, C=CH), 135.9 (C, C=CH), 135.8 (C, C=CH), 134.4 (C, C=CH), 132.0 (C, C=CH), 126.3 (CH, C=CH), 125.6 (CH, C=CH), 125.5 (CH, C=CH), 125.4 (2CH, C=CH), 120.1 (C, C2), 77.1 (CH, C4), 68.0 (CH, C5), 65.7 (CH2, C6), 40.9 (CH2), 40.8 (CH2), 40.7 (CH2), 35.7 (CH2), 33.9 (CH2), 29.2 (2CH2), 27.8 (CH2), 27.7 (CH2), 27.6 (CH2), 25.9 (CH3, C=C(CH3)2), 17.8 (CH3), 16.2 (2CH3), 16.1 (CH3) , 16.0 (CH3) ppm; SM (+APCI) m/z (%): 559.6 (100) [M + H]. The IR, 1H and 13C NMR spectra are presented in Supplementary Figs. S5, S6 and S7, respectively.
Preparation of the Vit C–SQ cream
Vit C–SQ (30.1 mg, 0.054 mmol) was weighted in a 5 mL vial. SQ (222 mg, 250 mL, 0.54 mmol) was added via syringe and the mixture was vortexed for 2 min. The obtained mixture was then sonicated using an ultrasonic cleaning bath to give a homogenous cream containing 3 mmol% of Vit C. A similar procedure was followed to obtain the Vit C–SQ cream containing 5 mmol% of Vit C.
Formulations for ex vivo skin testing
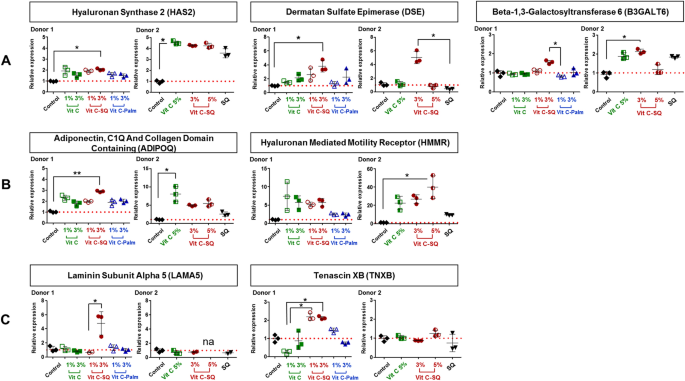
Explants from two donors aged 30 years were treated with the following formulations: (1) Vit C at 1 or 3 wt% in an aqueous cream of carboxymethyl cellulose (Sigma Aldrich) (2) Vit C–SQ conjugate dispersed in SQ; (3) Vit C–Palmitate complex; (4) squalenic acid in SQ (70:30, w:w) and (5) pure SQ. Final concentrations of Vit C in the various formulations were 1, 3 or 5 wt%.
Ex vivo activity testing on human skin
BIO-EC Laboratory possesses an authorization from the Bioethics group of the General Director Services of the French Research and Innovation Ministry (registered under n°DC-2008-542) to use human skin from surgical waste since 5th May 2010. The study was performed in accordance with the Declaration of Helsinki after the patients had given informed consent to use their skin samples by BIO-EC Laboratory.
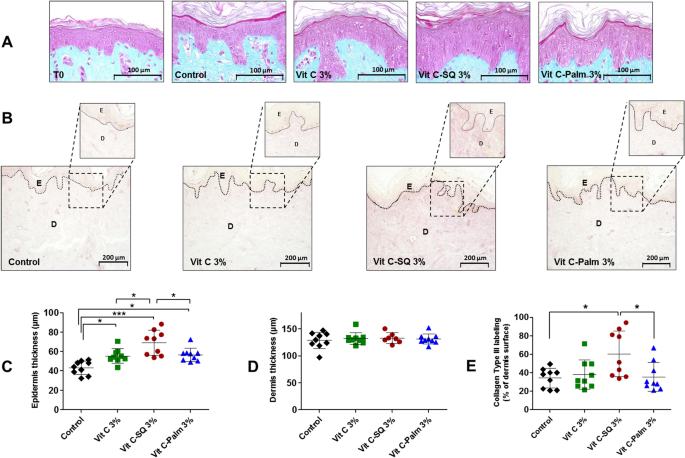
Skin explants (1-cm diameter) were recovered from an esthetic abdominal surgery in two 30-years old women and maintained in vitro for 9- or 10-days survival at 37 °C, in wet atmosphere enriched with 5% CO2, in BIO-EC's Explants Medium (BEM, BIO-EC Laboratory). At days 0, 1, 3, 6 and 8, explants received topical treatments with each cosmetic formulation (2 mg per explant) spread with a spatula, i.e. free Vit C, Vit C–SQ or Vit C–Palmitate at 1%, 3% or 5% (w:w). As a reference, pure SQ was also tested in one donor (donor 2). Untreated explants sampled at day 10 for donor 1 or 2 showed identical morphology as day-0 explants and were used as controls. Three explant replicates were prepared for each biological condition. Each skin explant was divided in two parts, one was fixed with ordinary Bouin and paraffin-embedded, the other one was frozen at − 80 °C until microdissection and genomic studies were performed.
Histological and immunohistochemistry studies
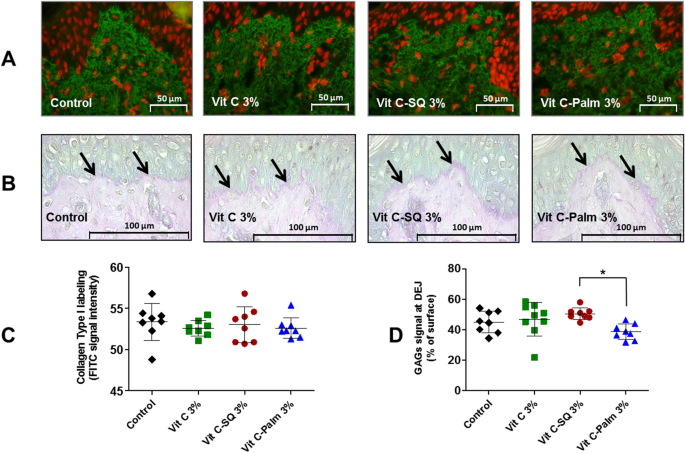
Five-5 µm tissue sections were prepared using a Minot RM 2125 microtome (Leica) and sticked on superfrost silanized glass slides. General tissue morphology was observed after Goldner-modified Masson trichrome staining. Glycosaminoglycans (GAGs) were examined on paraffin-embedded skin sections after staining with Schiff reagent. Neutral GAGs which link growth factors appear as purple pink near the DEJ. Optical microscopy observations were done with an Orthoplan microscope (Leica) at X40 magnification. Pictures were obtained with a tri CCD DXC 390P camera (Sony) and stored with Leica IM1000 software, version 1.10 (Leica). Collagen I immunostaining was done on frozen sections with 800-fold diluted polyclonal rabbit anti-human collagen I antibody (Monosan ref PS047) for one hour at room temperature, with a biotin/streptavidin amplification system, fluorescence revelation (FITC Caltag SA 1001) and nuclei staining with propidium iodide. Collagen III immunostaining was done on formol-fixed and paraffin-embedded slices with 50-fold diluted polyclonal goat anti-human collagen III antibody (SBA ref 1330-01), overnight at room temperature, with a Vectastain RTU Universal VECTOR avidin/biotin amplification system, DAB revelation.
Laser capture microdissection
Some skin explants were microdissected in order to separate dermis from epidermis. Serial sections (12-µm-thick human skin frozen tissue specimens) were placed on membrane slides (Carl Zeiss MicroImaging). Cresyl violet staining was performed by using a protocol from Zeiss Labs. Epidermal and dermal tissues were selectively dissected using the PALM MicroBeam system (Carl Zeiss). The dissection procedure was validated by measuring the differential expression of transcripts from genes specifically expressed in each tissue.
RNA extraction
Whole skin samples of 40–100 mg weight were homogenized in 1 mL TRI-Reagent (Euromedex), by agitation in a Precellys grinder with MK28-R metallic beads (Bertin) for two cycles of 3 × 20 s at 6500 rpm. After precipitation in 500 µL of isopropanol, RNA was high-speed centrifugated and the RNA pellet was washed twice with 75% ethanol, then solubilized in 30 µL of RNase-free water and stored at − 80 °C until molecular analyses. RNA purity and quantity were assessed by UV absorbance readings and RNA integrity was evaluated by capillary electrophoresis using RNA 6000 Nano chips and the 2100 Bioanalyzer (Agilent Technologies). RNA from microdissection samples were extracted in 300 µL TRI-Reagent, precipitated in 500 µL isopropanol overnight at − 20 °C, solubilized in 20 µL of RNase-free water and quality checked using Total RNA 6000 Pico chips and the 2100 Bioanalyzer. All RNA extracts showed homogeneous quality levels.
Amplification and reverse transcription of RNA
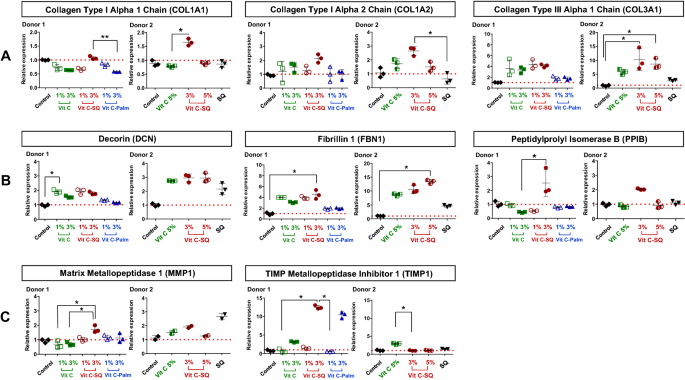
Prior to RT-qPCR study, 1 µg of total RNA from whole skin was amplified with the Quick Amp Labeling Kit (Agilent Technologies, ref 5190-0444), consisting of a double strand cDNA synthesis with MMLV-RT and oligo-dT-T7 promoter priming, followed by in vitro transcription with T7 RNA polymerase. Ten microliters of each amplified cRNA was directly reverse transcribed in 20-µL reactions, using Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics) and gene-specific priming with a 3-µL mix of qPCR primers designed for all genes studied (0.5 µM of each primer as final concentration in RT). The RNA-primers mix was incubated for 5 min at 65 °C, cooled on ice, completed to 20 µL with RT reagents and incubated for 30 min at 55 °C before enzyme inactivation for 5 min à 85 °C. Similarly, amplification of RNA extracted from each microdissected skin layer was performed from 10 µL of purified RNA solution and then RT was done with a priming mix at 0.07 µM final concentration in RT.
Real-time quantitative PCR
PCR primers obtained from the qPrimerDepot database55 or designed with Primer Designing Tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) were chosen overlapping an intron or hybridizing an exon junction when possible and checked for in silico specificity before synthesis by Eurofins Operon (Supplementary Table S1). qPCR analysis was done in triplicate 15-µL reactions with SYBR Premix Ex Taq Tli RNase H Plus mix (Takara) including 5 µL of 30-fold diluted cDNA and 0.5 µM of each primer. A 2-steps PCR program with hybridization at 61 °C followed by specificity check on fusion curves, was performed on CFX384 thermal cycler (Bio-Rad Laboratories). Quantitative values were normalized using the NORMA-Gene method56 for whole-skin data and with GAPDH and ACTB housekeeping genes for microdissection data57. Relative expression ratios were calculated by the delta-delta Cq method58, based on a theoretical efficiency considered at 100%, using as calibrator the untreated condition of similar duration as treated samples (day 10).
Statistical analysis
Data analysis was performed with GraphPad Prism version 5.0.0 for Windows (GraphPad Software, San Diego, California USA, www.graphpad.com). For epidermis and dermis thickness, collagen type I labelling and GAGs staining at DEJ, differences between treatments were tested using one-way ANOVA with Tukey's multiple comparison post-test, with variances homogeneity checking by Bartlett's test. For collagen type III labelling and gene expression fold-changes in whole skin explants, differences between treatments were tested using Kruskal–Wallis and post hoc Dunn's test. For expression data from microdissected skin, differences were tested by two-way ANOVA followed by Bonferroni's multiple comparison test. Differences between conditions were considered significant when p values < 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001). Quantitative data were graphically presented as mean ± standard deviation.